
Adrienne Loh (Physical & Biophysical Chemistry)
Over one-third of all secondary structures found in proteins are helices (1). It takes a special combination of amino acids, and usually a fair number of them (~20), to get a helix to form. However, peptides containing the unusual amino acid "Aib" (Figure 1) are known to form stable helices (2). This bias towards helical structures arises from the steric strain induced by the two methyl groups on the central carbon, and is strong enough that even very short peptides containing Aib are helical (3,4). Aib is therefore a useful amino acid for studying protein structure because it forces peptides to fold into helices.

Figure 1: "Aib" (a-aminoisobutyric acid). Aib is a post-translationally modified amino acid found naturally in bacteria.
It is also interesting to study helices that contain Aib because the particular type of helix adopted (there are several) and the stability of the helical structure is influenced by the length of the peptide, the amino acid sequence (if other types of amino acids are used) (5,6) and by peptide-solvent interactions (5-7).
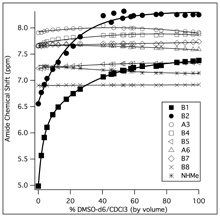
Undergraduate researchers in my group are currently investigating the stability and flexibility of Aib helices using NMR spectroscopy and circular dichroism (CD) spectroscopy. Helices are characterized by an intramolecular hydrogen-bonding network, where the amide proton on one amino acid forms a hydrogen-bond with an amide carbonyl on an amino acid that is three or four residues away. By studying the rate at which the amide protons exchange with deuterons from the solvent, we can learn something about the strength of the hydrogen-bond. If we measure these rates as a function of temperature, we can obtain activation energies for the exchange reaction. Thre presence of the hydrogen-bond can be identified by measuring the response of amide proton chemical shifts to changes in environment (solvent, temperature).
References:
1. Barlow, D.J. and J.M. Thornton (1988) J. Mol. Biol.
201, 601-619.
2. Toniolo, C., et al. (1985) Macromolecules 18, 895-902.
3. Paterson, Y., et al. (1981) J. Amer. Chem. Soc. 103, 2947-2955.
4. Prasad, B.V.V. and P. Balaram (1984) CRC Crit. Rev. Biochem. 16, 307-348.
5. Basu, G. and Kuki, A. (1992) Biopolymers 32, 61-71.
6. Pettijohn, A., Ph.D. Dissertation (1995), Cornell University.
7. Kennedy, D.F., et al. (1991) Biochemistry 30, 6541-6548.
Recently, students in my group have succesfully calculated the structures of four Aib-rich peptides with Ala, Lys, and/or Glu amino acids placed in different locations in the peptide helix. These calculations rely on NMR measurements of the interactions between nuclei: 1H-13C HSQC and 1H-13C HMBC spectra are used to assign chemical shifts to proton and carbon nuclei in the molecule by taking advantage of "through-bond" spin interactions. 1H-1H ROESY (similar to NOESY) spectra are used to determine how strongly nuclei interact "through-space", which provides information on the distances between nuclei that are not directly bonded (ie: close due to folding). This information is then used in combination with the freeware package Xplor-NIH to calculate the three-dimensional structure of the molecule. Manuscripts describing this work are currently in preparation.
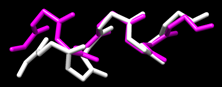
We have recently begun measurements of the interactions between our model peptide antibtioics and lipid systems (micelles, vesicles, etc.). Preliminary NMR evidence indicates that the peptides bind quite strongly, and that the peptides are well-structured. This is a new area for us, and research is in the early stages.
Loh Homepage UWL Homepage